Overview
Our research group is interested in understanding how RNAs change structures in order to perform their function. Until recently, only snapshots of molecules could be observed, hiding their modus operandi. We employ Nuclear Magnetic Resonance (NMR), and other biophysical techniques, to investigate the molecular mechanism of RNA function. When function of these molecular machines becomes apparent, it also provides a variety of unique new drug targets. The lab develops methods in NMR and RNA biochemistry to address these questions. Current projects include viral, bacterial and eukaryotic regulatory RNAs, e.g. microRNAs, ribosomal RNAs or RNA from HBV.
microRNA 34a
microRNA 34a
We study miR-34a, a microRNA that is dysregulated in various cancers and plays a central role in cellular and organism development. miR-34a targets many mRNAs coding for proteins important in diverse cellular functions (e.g. SIRT1, CD44, NOTCH, BCL2…). However, it is not fully understood how each specific mRNA is selected and when, among the myriad of mRNAs produced in the cell.
Understanding miRNA structure and motion will allow us to comprehend and manipulate miRNA function. The Petzold lab investigates the structure, dynamics, and thermodynamics of multiple miR-34a:mRNA complexes using NMR spectroscopy, molecular dynamic simulations, and a range of biophysical techniques. Recently, we discovered that miR-34a attains a high energy excited state (ES) conformation upon binding to SIRT1 mRNA (1). Stabilising this ES resulted in a two-fold increase in gene repression. Can miR-34a have a similar ES for other genes implicated in various cancers?
Revealing information about mRNA-specific conformations of miR-34a has profound therapeutic potential. Such conformations can further be stabilised by mutations to increase target specificity of miR-34a, leading to improved drug design and safer clinical prospectives. The Petzold lab is currently investigating such mutations that can increase targeting specificity.
In-cell NMR
In-cell NMR
NMR can offer atomic resolution of macromolecules in the cellular environment without requiring invasive probes, which can affect their function. However, most NMR studies of macromolecules consider them in buffered conditions. The cellular environment is inherently complex, containing different interaction partners, ionic strengths, and crowding. Our aim is to study nucleic acids in the cellular environment using NMR spectroscopy. Combining solution and solid state NMR we strive to measure dynamics of nucleic acids and push the boundaries of the size limitation by sensitivity enhanced methods. This will contribute to drug development (1).
Ribosomal RNA
Ribosomal RNA
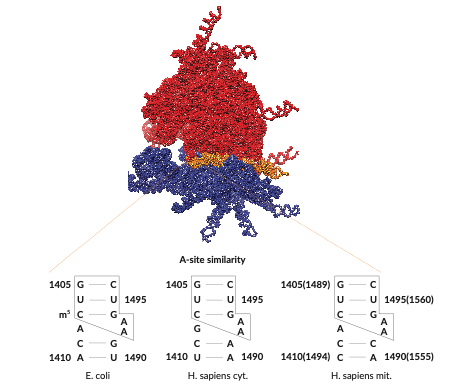
The ribosome is a complex RNA–protein nanomachine that translates messenger RNA (mRNA) into proteins in all living organisms. During the different steps of translation, the ribosome undergoes a number of global conformational rearrangements that have been studied in great detail. Local motions of individual ribosomal RNA (rRNA) helices, however, have received far less attention so far.1) These dynamics on the µs-ms timescale can influence interactions with certain classes of antibiotic agents and can potentially serve as actuators that drive the larger-scale rearrangements.2)
In our research, we study short-lived, structurally excited states in rRNA helices, which are not observable by conventional structure elucidation techniques. To this end, we investigate fragments of rRNA such as helix 44, the bridge between the small and large subunits, with NMR relaxation dispersion. By characterizing hidden excited states with atomic resolution, we can infer their impact on the structure of the ribosome. We then put our models to the test by investigating the relationship between excited state dynamics and ribosome function in living organism.
Our results will help increase our understanding of the similarities and differences between prokaryotic and eukaryotic ribosomes and expose a new regulatory layer – structural dynamics – that can be exploited for designing new antibiotic molecules with increased specificity against pathogenic bacteria.
Method development
Method development
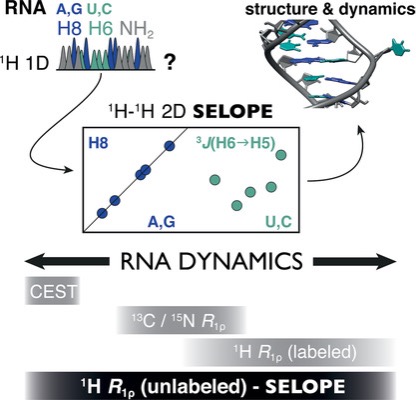
We are interested in studying the structure and dynamics of RNA using NMR spectroscopy. However, signal overlap and high cost of isotope labelling hinders this study. We strive to overcome these limitation by developing NMR methods to reduce signal overlap (1) which requires partial to no labelling (1H R1ρ). In addition, we are currently developing faster NMR acquisition techniques that can provide significant gain in time to study the structure and dynamics of nucleic acids. This can be used to study unstable RNA or RNA that is challenging to produce in high quantity. Our lab is also working in combining NMR with other methods (SHAPE, Cryo-EM etc.) to elucidate RNA structure and dynamics in greater detail.(2, 3, 4). Figure taken from (1).